# Past Projects
The embedded tweets take a bit to load! Sorry about that
# Queue-based clipboard with transcriber for easily capturing context for LLMs
peek at something I've been playing with
— Daniel George (@degtrdg) September 19, 2024
a queue-based clipboard with a built-in transcriber.
it's brought the activation energy of asking good questions to claude down to zero because it captures my thought process as i'm thinking. i honestly hate having to context switch… pic.twitter.com/GjFBxjiBul
- tool that brings the activation energy of asking good questions to claude down to zero because it captures my thought process as i’m thinking
- reduces context switch between reading, copypasting, typing, etc while focusing
# AI-enabled chart generation
Scientists should spend more time doing science and less time fighting with their software.
— Nicholas Larus-Stone (@nlarusstone) September 4, 2024
Introducing @sphinx_bio’s new AI-enabled cell editing, a way for scientists to answer their most important questions in a fraction of the time!
Now you can simply tell Sphinx what you’re… pic.twitter.com/Pff8P9wuSz
- I spearheaded this feature from ideation, implementation, and set the foundation for Sphinx’s LLM evaluations with what I made here
- Read more details in the blog: https://www.sphinxbio.com/post/ai-enabled-chart-creation
# PreScribe
Demo
An AI medical scribe that proactively checks off requirements for medicine as the conversation goes on, real-time.
# Proposal Reviewer
Demo
Creates a diff on your writing given the weakness of your writing. Only looks at the most relevant parts of your writing and gives specific deletions and inserts to the writing to address the weakness.
Links:
# Personalized Discovery Fiction Generator
Demo
Using LLMs to create discovery fiction (like https://michaelnotebook.com/df/index.html) where the model creates a first-person narrative of the information of how the student could have discovered what they’re learning.
# LLM-based Gene Perturbation Simulator
LLM-based Gene Perturbation Simulator
— Daniel George (@degtrdg) March 17, 2024
Put in a target gene, what you did to the gene, and what phenotype you're looking for, and simulate your perturbation on the rest of the gene network! pic.twitter.com/zQ1rx09nUl
# Perceptual filter
Perceptual filter based on what's important to you!
— Daniel George (@degtrdg) January 19, 2024
Made this w/ @ahadj0 on having an LLM perceive what you will see and only show it to you if it's worth your time.
You define what's important to you and what's not. It reasons whether a tweet in your feed should show up! 1/n pic.twitter.com/02GdxyBmao
# The Orwell Editor v1
Introducing The Orwell Editor v2 ✨
— Daniel George (@degtrdg) April 30, 2023
Now there is:
- copy & paste for your writing to have it analyzed
- shortcuts to make post-it notes
- I'm trying more non-linear writing, so lmk how it is
- upgraded UI to be unobtrusive
Link is below.
cc: @_buildspace @_nightsweekends pic.twitter.com/nqUeRBhVyc
# Tools for Stepping into Biology
What would a more human medium for sharing understanding in biology look like? Mediums that shed the assumptions of our limitations from when paper was SOTA? https://t.co/Osq41k0iZH
— Daniel George (@degtrdg) October 14, 2023
Thanks to @NikoMcCarty for getting me to publish my first piece! pic.twitter.com/l0jPDe0X2k
# Synapse: Insights in context of when you need them
Demo
Writing interface that brings up relevant but slightly tangential work in a reduced representation to give peripheral vision ( https://notes.andymatuschak.org/Peripheral_vision) on things that might lead to a new direction of work. Data is sourced from a the researcher’s citation manager (Zotero). Researchers accumulate a pristine dataset of what’s most import in these citation managers which can lead to interesting things coming up. The reduced representation of the source gives the user the gist of the idea that they’re already familiar with.
# Whisper Notes
Demo
A simple voice-to-text clipboard app for quickly recording your thoughts. Use global commands to record and fill your copy register with the transcript. View and delete previous transcripts.
Links:
# BioConceptVecXplorer
Demo
Knowledge is continuous, but its representation in papers is discrete. We can instead represent concepts as vectors and explore a more fluid latent space. Using vector embeddings trained on 30 million PubMed abstracts, we created a tool for researchers to create biological analogies to discover relationships not explicitly in the literature. This was previously done in material science to discover an anti-ferromagnetic material not explicitly in the literature. We extended this idea as a tool to enable bioengineers to make discoveries as well.
We were interviewed and written about for this project on the following biotech newsletter .
Links:
The following are mostly from the genetics lab I work at
# Spheroid Analysis
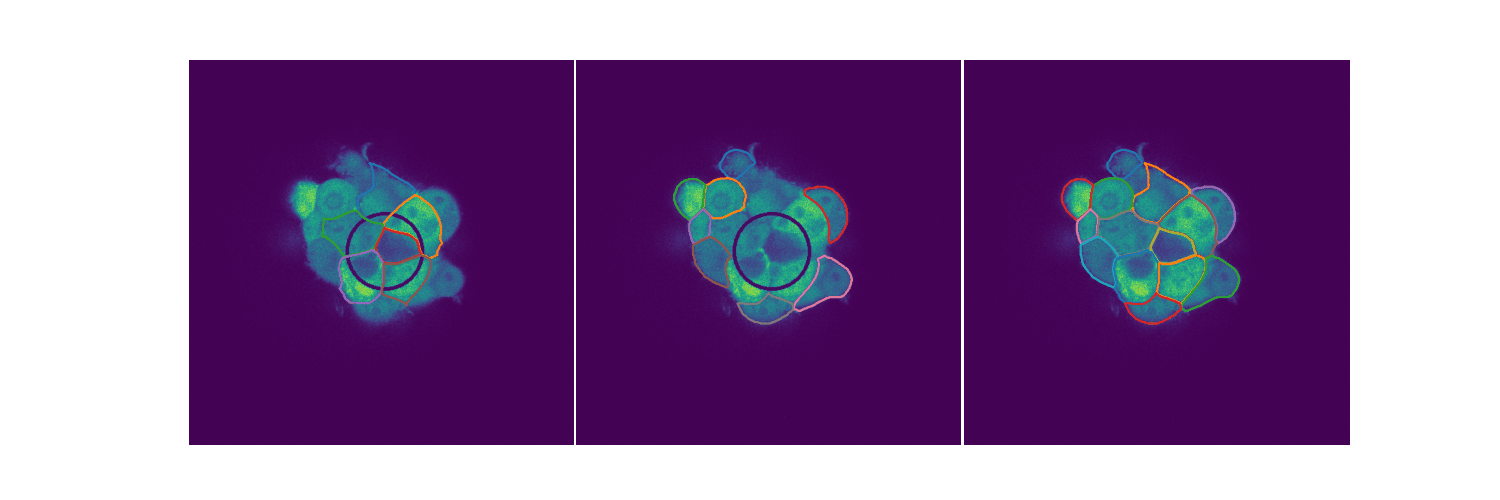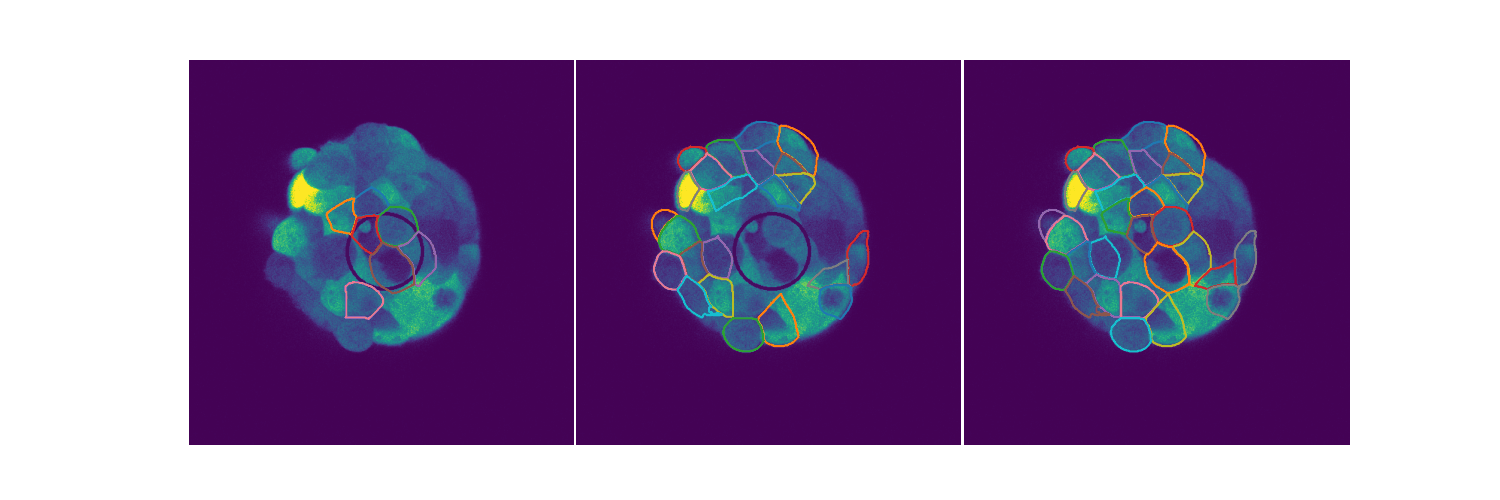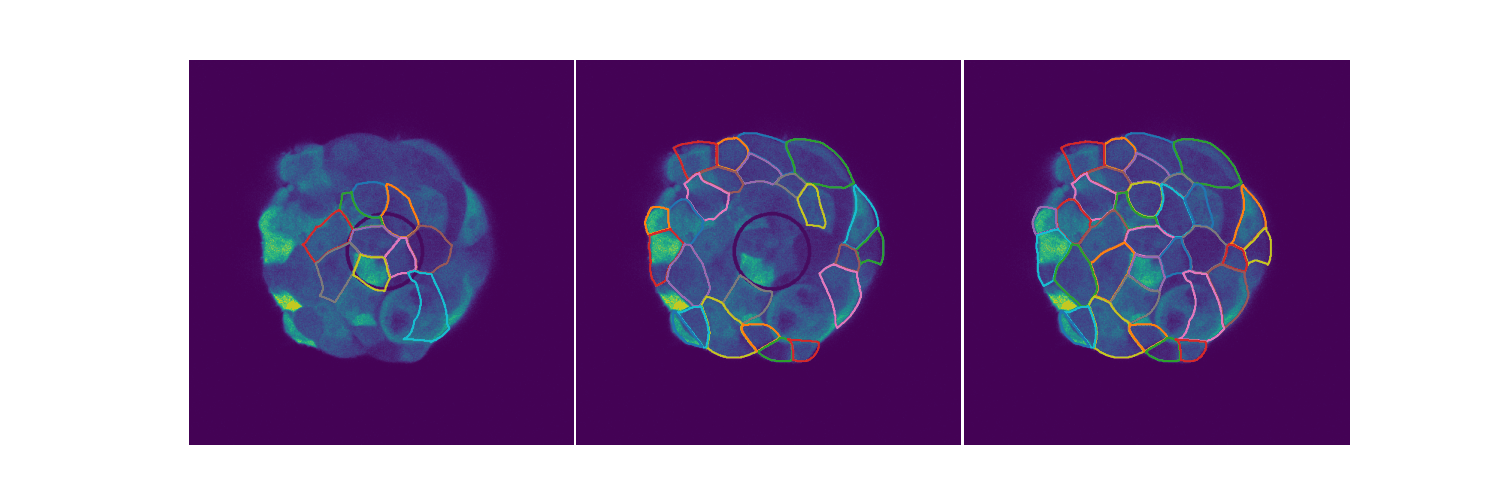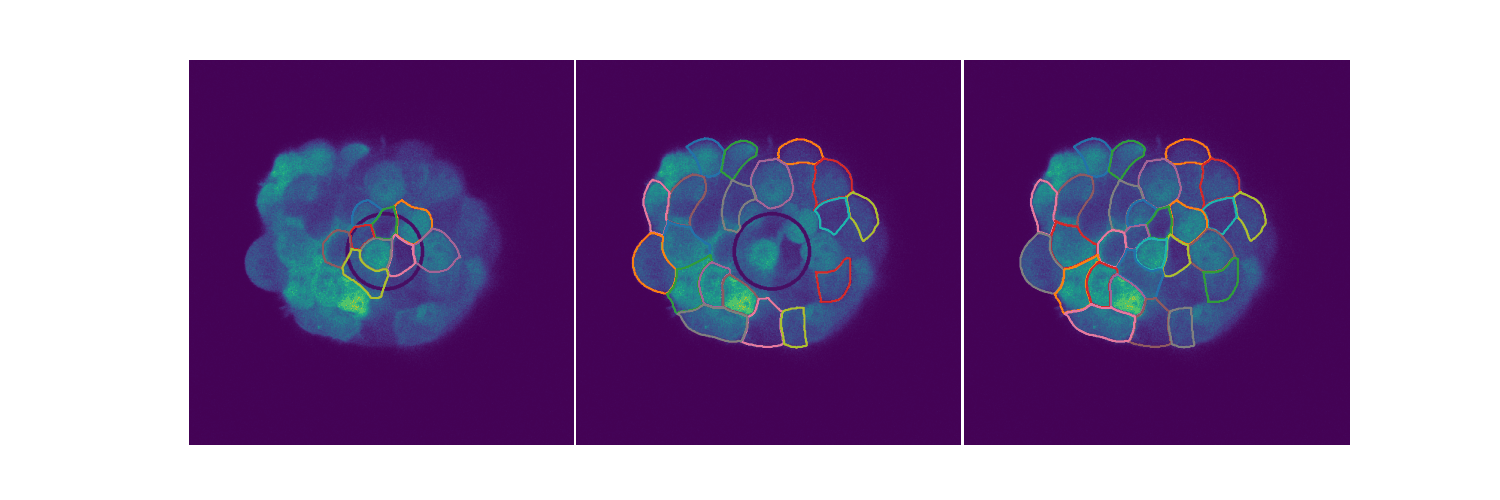
Example plot from one of my EDAs
Spheroids are 3D cultures that mimic tissues and micro tumors better than the 2D cultures we see in Petri dishes for a variety of reasons. When extending biological circuits from the 2D to the more realistic 3D, the morphology of the cells could affect things like protein production. Protein synthesis is an inherently stochastic process, so when creating biological circuits quantifying a measure of noise in the system is quite useful. This is extending the 2D analysis to spheroids. You can find out more in my slides.
Links:
# Physically Unclonable Function (PUF) Pipeline and Analysis
PUFs are a type of fingerprint that is introduced in manufacturing to show the lineage of a device. They are mainly used in integrated circuits, but they can also be used in other domains like biology. Using CRISPR one can make a PUF to that is impossible to reproduce and can be used to determine the lineage of a cell line ( CRISPR-PUF). I wrote a pipeline to process the millions of DNA sequencing reads that are used to find a ‘distance’ between cell lines. If they are the same cell line the distance would be small, and it would be large if the cell lines are different.
Links:
# Finding Genomic Safe Harbors for CHO
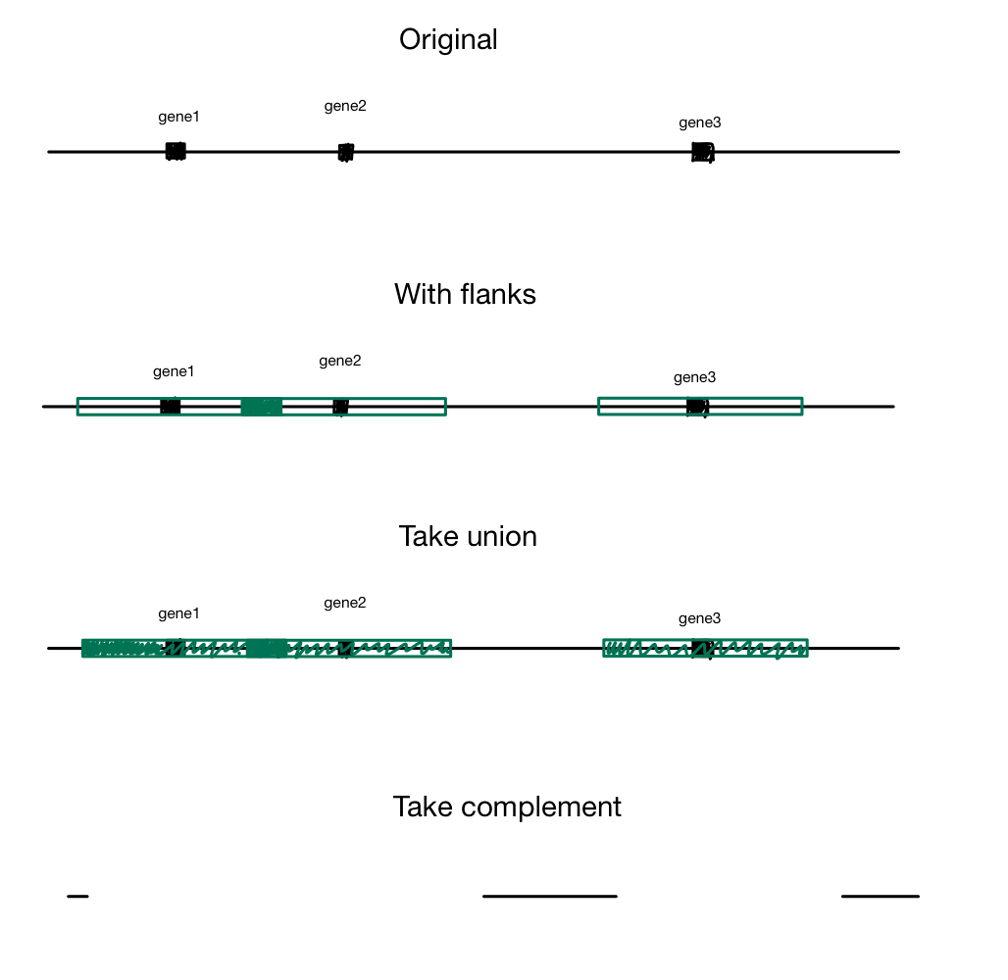
Logic behind pipeline from slides
When you edit a cell line with techniques like CRISPR, if you add code without taking into account the function of the regions around it, there might be unintended consequences. There are certain regions that are considered safe harbors where your changes won’t interfere with existing functionality. There was code from a paper which defined requirements for safe harbors for the human genome, but we needed it to work for the Chinese Hamster Ovary, so I wrote a Python port than can be extended to any genome. See slides for more info.
Links:
# Projects
# Small Molecule Autocomplete RNN
I trained a LSTM based RNN to efficiently enumerate chemical space given a SMILES input. It uses BFS to take the top few possibilities in the probability distribution, and I made model more creative and incentivized it to give shorter outputs.
Links: